There are many risks associated with trusting a poorly written structural alert; an incorrect prediction could result in wasted time and money. At Lhasa Limited, we take pride in the accuracy and consistency of our products. This article aims to elucidate alert reliability and discuss the process of expert driven alert creation in Derek Nexus.
Derek Nexus contains alerts for multiple endpoints, including carcinogenicity, mutagenicity, genotoxicity and skin sensitisation. These alerts can predict the potential toxicity of a compound, but how are they created?
Alert creation in Derek Nexus
Extensive research is carried out using public and, where available, proprietary data sources. The evidence is assessed using defined criteria to identify the key data and knowledge that can be used. Where a toxicophore is identified, the Structure-Activity Relationship (SAR) is refined using available data to define the boundaries for an alert. The scope of the alert is implemented as a “pattern” to which query compounds will be compared to, and this is also summarised in a graphical representation for the user in an “alert description image”. Reasoning rules are used to reflect the probability of the toxicity occurring and modify the outcome depending on species type, biological and / or physiochemical properties. The scope of the alert is defined by taking into consideration the availability of detailed study data, metabolic activation, example structures and proposed mechanisms of toxicity. The alert is then implemented into a knowledge base using Derek Knowledge Editor. When compounds are processed against the knowledge base in Derek Nexus, a toxicity prediction is generated for those query compounds that match the scope of the alert.
What about proprietary data?
When considering proprietary data, we ask members to provide as much detail as possible (e.g. metabolic activation, strain profile and dose response data) for analysis. This helps us to refine the SAR and further support any potential mechanisms for the toxicity. Where there is only high-level data available to us, for example positive / negative summary calls per compound, we are still happy to consider this – there is an element of trust that the study will have been carried out to comply with relevant guidelines and summary calls have been made appropriately following analysis by the expert internally.
From a member’s perspective, it can sometimes be difficult to understand where these alerts emanate and know they can be trusted. For an individual member who donates proprietary data, we check any alerts derived from the data and obtain approval prior to incorporation into the general knowledge base to ensure confidentiality. For the wider membership we have established a role as trusted partners when it comes to extracting general alerts from proprietary data, through industry wide collaboration.
What makes Derek Nexus’ knowledge base trustworthy?
Data in Derek’s knowledge base is considered on a case-by-case basis by assessing the quality of the study. The decision to accept / reject data relies on:
- Expert review
The experimental method and data are reviewed by the scientist implementing the alert to ensure consistency, for instance for the Ames test we look for a dose-dependent doubling of revertant counts at an appropriate concentration, alongside adequate control data. All work is checked twice; first the individual alert is peer reviewed by a colleague, and then again after integration into the main knowledge base by the project team.
- Reliability
Any deviations from standard protocol that may affect the reliability of a study is taken into consideration and, if considered appropriate to include within the alert, this information will be transparently described in the alert description, including references to studies considered.
Accuracy of Derek Nexus
The validation feature within alerts for several endpoints provides the user with information about the predictive performance of an alert against one or more data sets, which may be publicly available or proprietary data. The user can consider this information to assess the reliability of each individual alert which may increase confidence in the prediction.
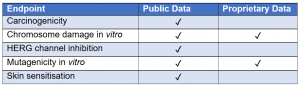
Figure 1. Table to show which endpoints are validated as public data or proprietary data in Derek Nexus.
The predictive performance of Derek Nexus may vary depending on the application of interest (e.g. the predictivity for mutagenicity differs from drug impurities to APIs). However, at the bottom of this article is a list of relevant references that illustrate the performance for the mutagenicity endpoint in Derek Nexus for various use cases.
Additionally, a recent publication ‘Evaluation of the Global Performance of Eight in silico Skin Sensitization Models Using Human Data’, evaluates the accuracy of eight in silico skin sensitization models against two human data sets. Derek Nexus performed exemplary, achieving the highest score for correctly predicted and the lowest combined score for incorrect / no prediction. Our recent infographic, ‘Derek Nexus – Achieving High Accuracy with High Coverage’ summarises these findings.
The Derek Nexus model is described and documented according to the OECD standard using the QSAR Model Reporting Format (QMRF) for the mutagenicity, chromosome damage, carcinogenicity, skin sensitisation, teratogenicity and HERG channel inhibition endpoints. QMRF documents may be used to help support regulatory assessment of chemicals and current versions of QMRF documents are available on our website.
In conclusion, alerts in Derek’s knowledge base are created by expert scientists who assess the quality of available study data on a case-by-case basis to ensure reliability. Reliability and applicability of an alert activated for a particular query compound can be further assessed by the user by considering the alert description image and comments which transparently detail the relevant information, supporting references and rationale used to derive the alert.
Visit our website to find out more about Derek Nexus or get in touch with the Business Development Team for a free demo.
Relevant references
| Industry | Mutagenicity reference |
| Pharmaceuticals | Snyder, R.D. (2009). ‘An update on the genotoxicity and carcinogenicity of marketed pharmaceuticals with reference to in silico predictivity’. Environ. Mol. Mutagen., 50, 435-450. https://doi.org/10.1002/em.20485 |
| Pharmaceutical Impurities | Dobo, K., et al. (2012). ‘In silico methods combined with expert knowledge rule out mutagenic potential of pharmaceutical impurities: An industry survey’. Regul. Toxicol. and Pharmacol., 62(3), 449-455. https://doi.org/10.1016/j.yrtph.2012.01.007Williams, R. V., et al. (2016). ‘It’s difficult, but important, to make negative predictions’. Regul. Toxicol. and Pharmacol., 76, 79-86. https://doi.org/10.1016/j.yrtph.2016.01.008 |
| Occupational Health | Araya, S., et al. (2015). ‘Mutagenicity assessment strategy for pharmaceutical intermediates to aid limit setting for occupational exposure’. Regul. Toxicol. and Pharmacol., 72(2), 515-520. https://doi.org/10.1016/j.yrtph.2015.10.001 |
| Flavour Chemicals | Ono, A., et al. (2012). ‘Validation of the (Q)SAR combination approach for mutagenicity prediction of flavor chemicals’. Food Chem. Toxicol., 50(5), 1538-1546. https://doi.org/10.1016/j.fct.2012.02.009 |
| Industrial Chemicals | Hayashi, M., et al. (2005). ‘In silico assessment of chemical mutagenesis in comparison with results of Salmonella microsome assay on 909 chemicals’. Mutat. Res., 588(2), 129-135. https://doi.org/10.1016/j.mrgentox.2005.09.009 |
| Pesticides | Worth, A., et al. (2010). ‘The Applicability of Software Tools for Genotoxicity and Carcinogenicity Prediction: Case Studies relevant to the Assessment of Pesticides’. JRC Scientific and Technical Reports. https://www.doi.org/10.2788/61485 |
| Cosmetics | Ates, G., et al. (2016). ‘In silico tools and transcriptomics analyses in the mutagenicity assessment of cosmetic ingredients: a proof-of-principle on how to add weight to the evidence’. Mutagen., 31(4), 453-461. https://doi.org/10.1093/mutage/gew008Golden, E., et al. (2021). ‘Evaluation of the global performance of eight in silico skin sensitization models using human data’. Altex., 38(1), 33-48. https://doi.org/10.14573/altex.1911261 |
| Food / Food Ingredients | Mertens, B., et al. (2017). ‘Coatings in food contact materials: Potential source of genotoxic contaminants?’. Food Chem. Toxicol., 106(A), 496-505. https://doi.org/10.1016/j.fct.2017.05.071 |
| Food Contact Materials | Vuorinen, A., et al. (2017). ‘Applicability of in silico genotoxicity models on food and feed ingredients’. Regul. Toxicol. and Pharmacol., 90, 277-288. https://doi.org/10.1016/j.yrtph.2017.09.026 |
| General | Myden, A., et al. (2017). ‘Utility of published DNA reactivity alerts’. Regul. Toxicol. and Pharmacol., 88, 77-86. https://doi.org/10.1016/j.yrtph.2017.05.016 |
| QMRF | QMRF report for Derek Nexus mutagenicityAll of Lhasa’s QMRF Reports |
Last Updated on January 25, 2024 by lhasalimited



