It is common knowledge to those submitting drug substances for regulatory assessment, that an in silico assessment of mutagenic potential may be conducted in lieu of carcinogenicity or bacterial mutagenicity data for any impurities. A key requirement in ICH M7 states that “Two (Q)SAR prediction methodologies that complement each other should be applied. One methodology should be expert rule-based & the second should be statistical-based“. Combining predictions utilises the advantages of each system, compensating for the limitations of either, providing a more sensitive and conservative assessment that is essential for identification of human health hazards. However, this can occasionally result in conflicting predictions therefore it is important to understand each system to undertake expert review and conclude a single result.[1,2]
Numerous free and commercial (Q)SAR tools are available to complete a computational assessment of mutagenicity. Although it is a requirement of ICH M7 to use two complementary (Q)SAR methodologies, the assessor is free to explore the strengths and limitations of all (Q)SAR systems – which “follow the general validation principles set forth by the Organisation for Economic Co-operation and Development (OECD)” – as it is not essential to use those provided by a single vendor. Several questions must be answered when generating predictions using multiple systems:
- How do I combine these two predictions to provide a single conclusion?
- How do I ensure I have all the necessary information from both systems?
- How do I know which system to trust?
- How do I prepare different outputs as a single report?
- How do I store the information for future reference?
Derek Nexus and Sarah Nexus, Lhasa Limited’s expert rule-based & statistical-based systems respectively, were recently released in Nexus. Leveraging the advantages of both being contained in the Nexus platform, it is worth recognising some of the benefits provided when systems can be integrated to provide a full assessment in one environment.
Here are 5 examples of how using Derek and Sarah in tandem can make mutagenicity assessment a simpler task:
Combines two predictions to a single ICH M7 prediction
Nexus avoids the need to compile results through use of a combined ICH M7 prediction. A single click generates both the Derek mutagenicity in vitro and Sarah predictions as well as a single “In Silico Overall Call”.
Includes a guided review process
An automated “In Silico Expert Review” feature was implemented in Nexus 2.3. When an ICH M7 prediction has run, the user is presented with compound-relevant standardised expert review arguments depending on the prediction scenario. Although the final decision lies with the assessor, these arguments guide the user by highlighting what information in the Derek and Sarah predictions is expected to require review to conclude a single prediction.
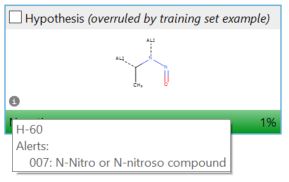
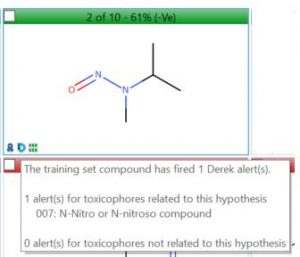
These automated arguments lead the reviewer to the relevant information but it can still be challenging to marry the SAR of the impurity with the Derek alert pattern(s) and any Sarah hypotheses with training examples. Therefore, a label is attached to any Sarah hypothesis and training set example highlighting it’s associated Derek alert(s).
 |
 |
Highlights additional information
Sarah contains a heat map showing which strains in a compound have been tested. It is simple to check whether the compound has been tested in the 5 strains recommended by OECD TG 471 or the strain considered most likely to show activity.
Lhasa have undertaken significant research on nitrosamines this past year,[3] refining the Derek alert to restrict certain nitrosamines considered to be inactive in the Ames test. Although scientifically justified, nitrosamines, as chemicals in a cohort of concern, require to undergo assessment on an individual chemical basis. Therefore, within the latest release of Nexus 2.4, arguments are now automatically displayed for any query considered to belong to a cohort of concern, emphasising that a compound specific risk assessment must be made regardless of the prediction.
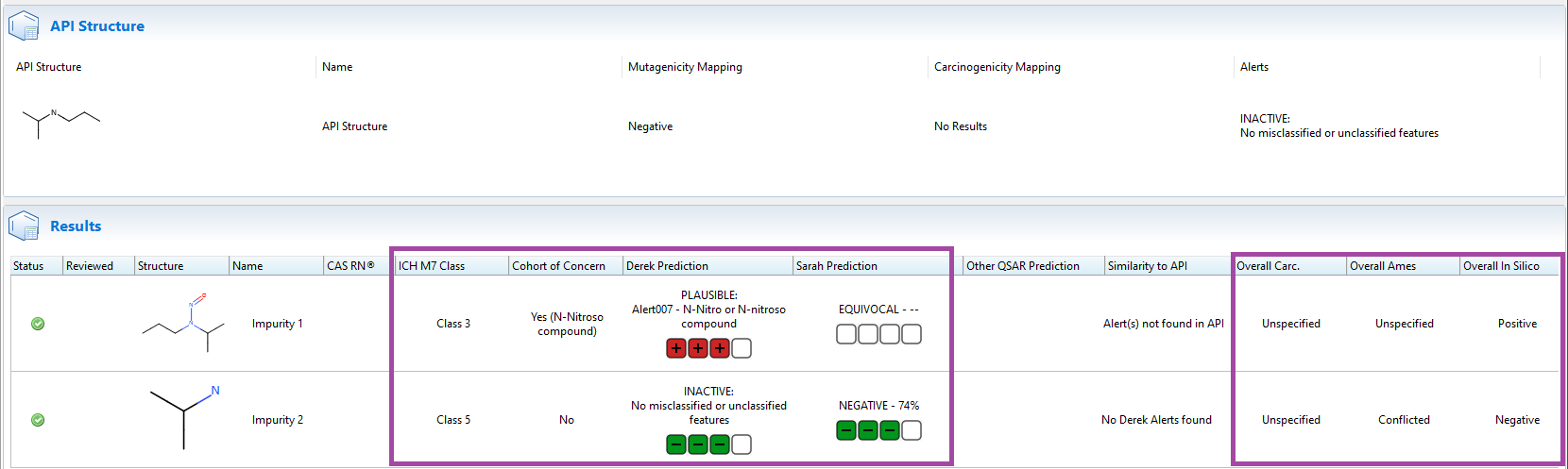
Provides a full ICH classification
The primary use case for (Q)SAR predictions under ICH M7 is to assess the Ames mutagenicity of all potential impurities alongside the API. The Nexus ICH M7 classification tool processes known impurities alongside the API to generate a predicted ICH M7 class for each impurity based on the in silico predictions as well as the known mutagenicity and carcinogenicity of the API and impurities. This feature also benefits from Ames and carcinogenicity data leveraged from Vitic & the Lhasa Carcinogenicity Database.
 |
Data storage
Smart storage of (Q)SAR assessments is essential so that previously reviewed compounds are readily accessible & sharable. As a result, Setaria is a knowledge-searchable database for the storage of toxicity data to manage impurity assessments under ICH M7. It benefits from cohesive integration with Derek & Sarah, allowing predictions to be pulled through from Nexus.
If you are interested in learning more about any of the in silico tools mentioned in the article, please get in touch with our Applied Sciences Team to arrange a free trial or demonstration.
References
- Foster et al. (2020) ‘The importance of expert review to clarify ambiguous situations for (Q)SAR predictions under ICH M7’, Genes and Environment, 42(27). https://www.dx.doi.org/10.1186/s41021-020-00166-y
- Williams et al. (2015) ’Establishing best practise in the application of expert review of mutagenicity under ICH M7’, Regulatory Toxicology and Pharmacology, 73(1), 367-377. https://www.dx.doi.org/10.1016/j.yrtph.2015.07.018
- Thresher et al. (2021) ‘Are all nitrosamines concerning? A review of mutagenicity and carcinogenicity data’, Regulatory Toxicology and Pharmacology, 116, 104749. https://www.dx.doi.org/10.1016/j.yrtph.2020.104749
Last Updated on January 25, 2024 by lhasalimited